Compare multiple of spectra
Multiple spectra analysis allows to quickly extract information about a set of spectra.
Spectra selection, normalization and previsualization
The first step is to select the spectra :
How to select spectra.
Spectra selection
All the spectra analysis tools start with a phase of selection.
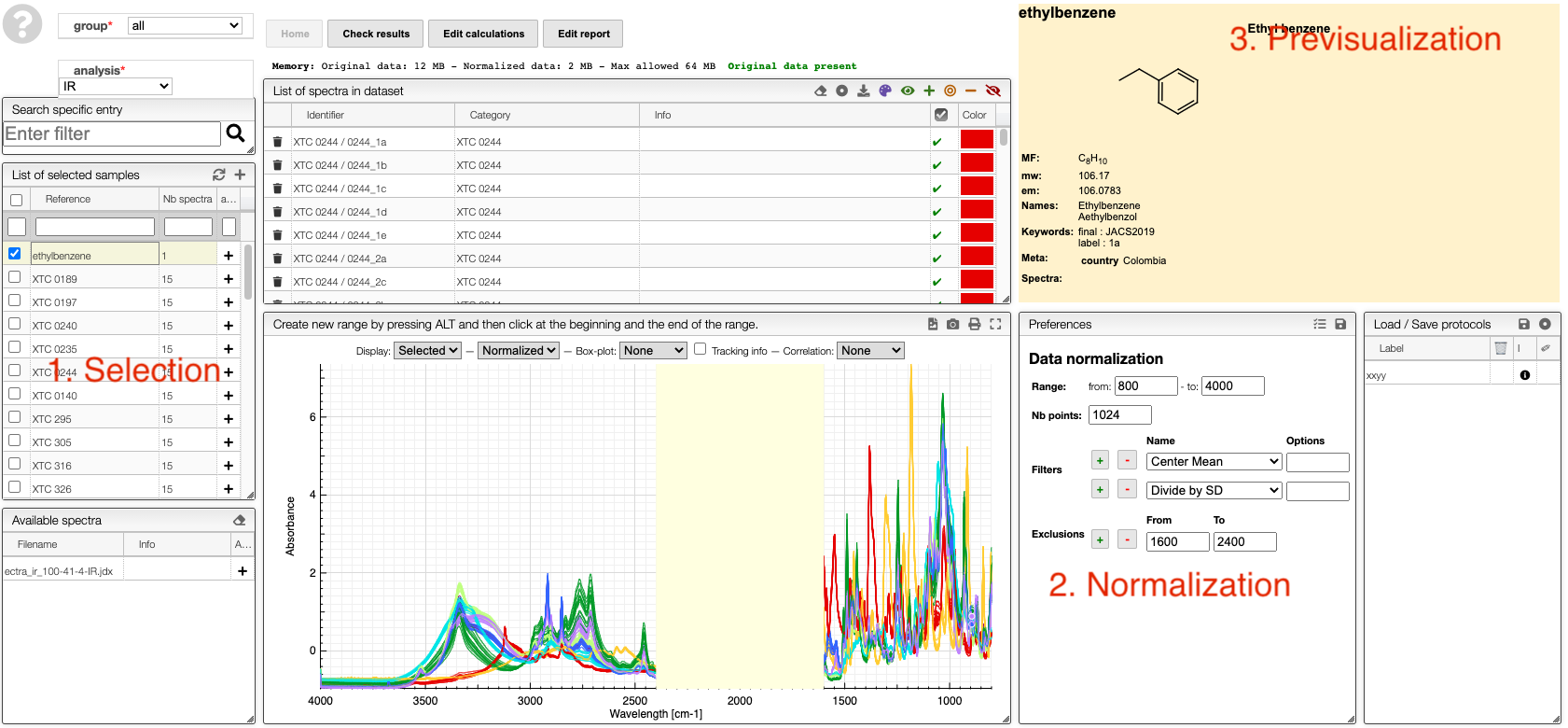
Select samples
In order to facilitate the analysis of the spectra it is advised to have samples containing representative spectra in order to evaluate the intra-variability as well as the reproducibility.
Selection of spectra to analyze is achieved with one of those 3 methods:
At the level of the sample by either clicking on the +, this will add all the spectra related to this sample or on the + on the top of the sample box to add all the spectra of all the selected samples.
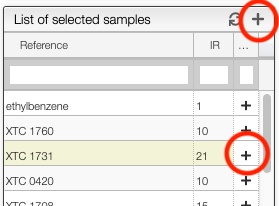
If you select a sample it is also possible to add a specific spectrum by clicking on the + at the level of the spectra list.
Once spectra have been selected, data normalization filters can be applied :
Apply mathematical tools to the spectra.
Preprocessing
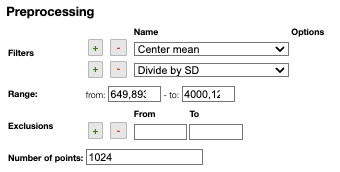
Filters
You can apply the following filters to the spectra to enhance the visualization. The modifications include the following:
Center Mean: subtract the mean from every variable observation in the dataset, so that the new variable's mean is centered at 0.Center Median: subtract the median from every variable observation in the dataset, so that the new variable's median is centered at 0`Divide by SD: divide every variable observable in the dataset by the standard deviation yields a distribution with a standard deviation equal to 1.Normed: Specify a value in thevaluefield and select the type of normalization:Sum to value: normalize the integral under the curve so that it sums to the specified value.Absolute sum to value: normalize the integral under the curve so that the absolute sum sums to the specified value.Max to value: normalize the maximum value to the specified value.
Rescale (x to y): rescale the graph such that the y-values fit between specified minimum and maximum values.First derivative: calculate the first derivative of the spectra.Second derivative: calculate the second derivative of the spectra.Third derivative: calculate the third derivative of the spectra.Savitzky-Golay: smooth the spectra and calculate derivatives based on the following parameters:Window: smoothing window size, must be an odd number, greater than 5.Derivative: derivative order.Polynomial: the degree of the polynomial used to calculate the Savitzky-Golay.
AirPLS baseline: baseline correction using adaptive iterative reweighed penalized least squares algorithm.Iterative polynomial baseline: baseline correction using iterative polynomial fitting algorithm.Rolling average baseline: baseline correction using a rolling average.Rolling median baseline: baseline correction using a rolling median.Rolling ball baseline: baseline correction using a rolling ball.Ensure growing X values: ensure that the x-values are in increasing order.Function on X: apply a function to the x-values. For example,log(x).Function on Y: apply a function to the y-values. For example,log10(y+1).Calibrate X: calibrate the x-values with the parametersfrom,to,nbPeakandtargetX.Pareto normalization: Pareto scaling, which uses the square root of standard deviation as the scaling factor, circumvents the amplification of noise by retaining a small portion of magnitude information. 10.1016/j.molstruc.2007.12.026
One classical preprocessing algorithm is Standard Normal Variate (SNV). This preprocessing can be achieved by selecting the 2 options Center mean and Divide by SD.
Selecting the range
A certain range of x-values can be selected to show only a part of the spectrum using Range.
Exclusions
Depending on the analysis, some regions should be removed using Exclusions in order to improve the visualization.
Number of points
Number of points can be changed to reduce the number of points in the spectra.
The superimposed spectra can be manipulated without numerous :
How to visualize spectra.
Spectra visualization
Numerous options are available to display the either all the spectra in the dataset or the selected spectra in the dataset.
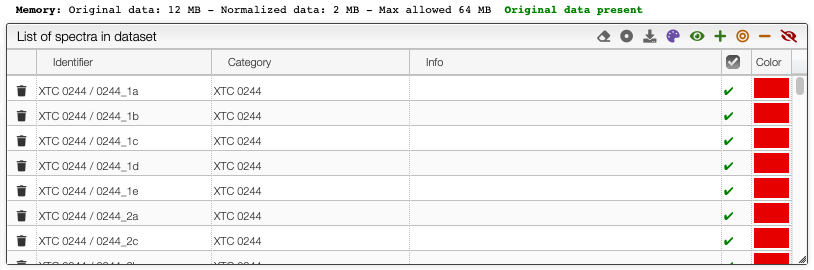
Selection of spectra in the dataset
The toolbar on the top of the list of spectra in the dataset provides many options (from left to right):
- Remove all spectra from dataset
- Select category: select which property contains the category description
- Download normalized matrix
- Recolor spectra based on category: a different color will be applied for each category. By default, the sample reference
- Select all spectra
- Append to selected spectra
- Select only current spectra
- Remove spectra from current selection
- Unselect all spectra
Graph options
It is possible to either display the selected spectra, all the spectra or various derived information.
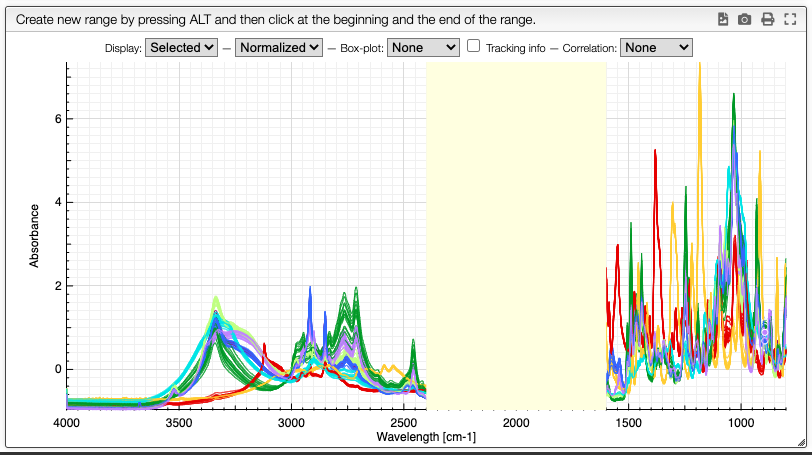
Customization of the display is achieved using the chart toolbar:
Display spectra
The first options allow to either display all the spectra, only the selected spectra or nothing.
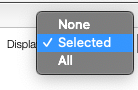
Displaying no spectrum is useful when displaying other derived data.
Original / normalized
These options allow to either display the original spectra or the normalized data. Most of the time we will display normalized data. Those are the data that will be analyzed, and normally they also take less room in memory.

Boxplot
The boxplot kind of representation allows to display the first / third quartile as a dark grey zone for each X point. The min and max values are represented as a light gray zone and the median is represented as a line for which the color varies based on the standard deviation (red: high variation, blue: small variation).
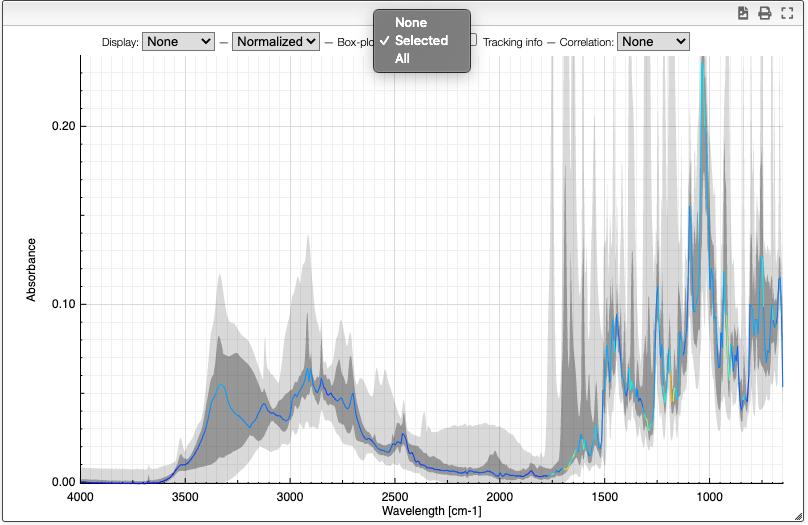
Tracking information
By selecting the tracking information you will display the X values and the corresponding Y values for all the spectra.
![]()
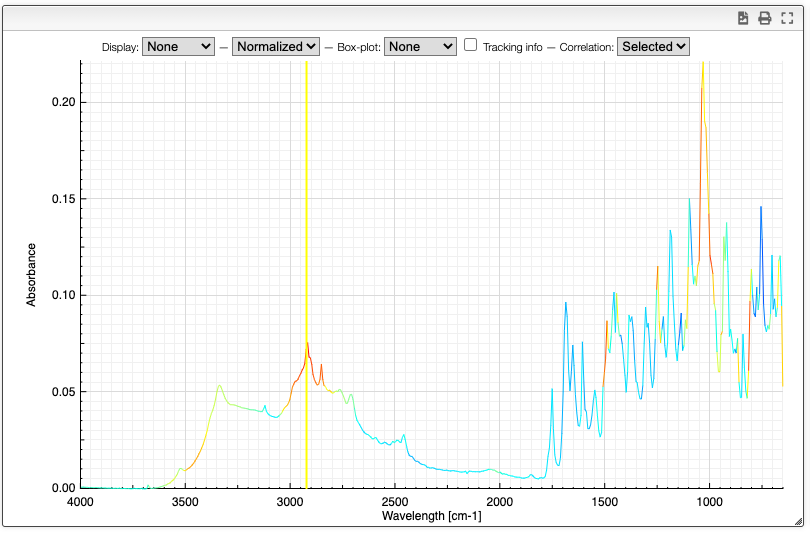
Correlation
Correlation of the vector represented by the Y points can be useful to determine which peaks are correlated in a big mixture of products. This is known in NMR metabolomics as STOCSY.
By SHIFT ⇧ + ALT + click you can select the X value for which you would like to check correlation. Strongly correlated signals will appear in red while non correlated signals are blue.
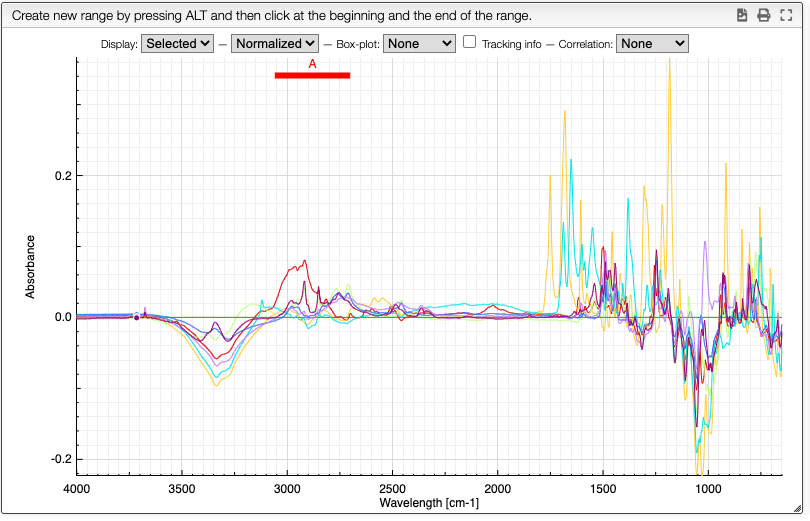
Comparing spectra
Once you have superimposed spectra you can define ranges by maintaining alt pressed and click once left and right of the range you want to define.
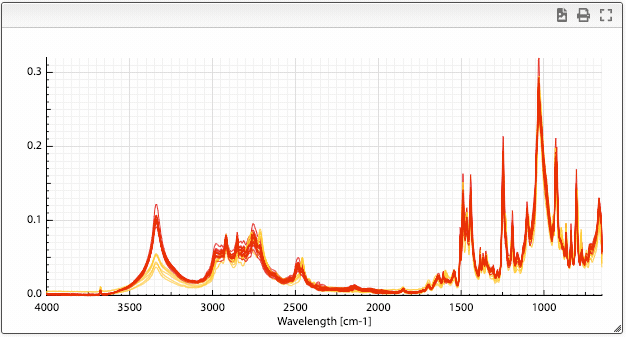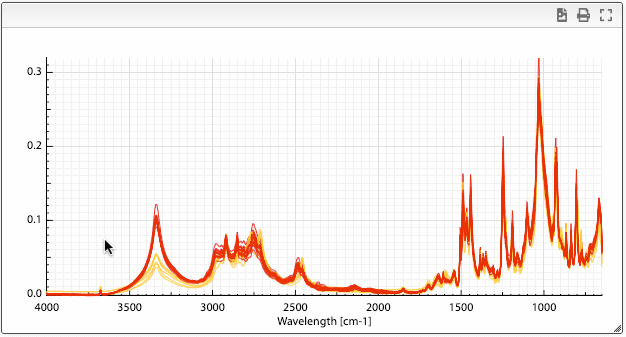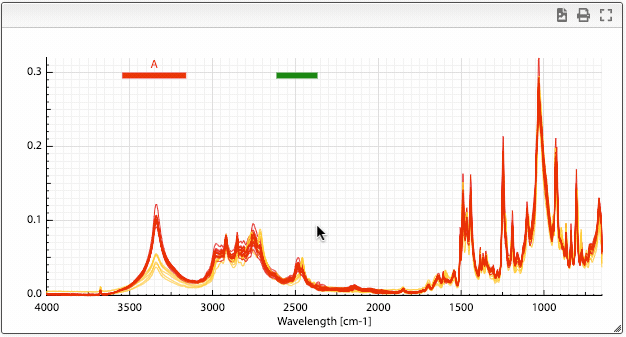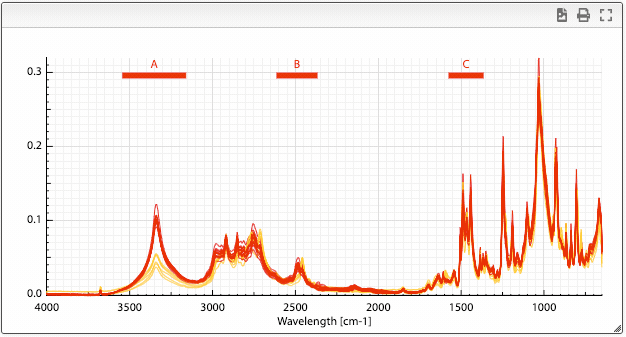
A column will be added in the dataset table that contains the integration of the selected area. It is also possible to define more than one range by repeating this procedure.
In the Define ranges tab it is possible to rename the variables as well as to define custom formulae based on the ranges integration.
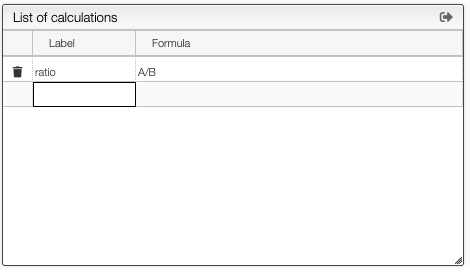
All the ranges and custom calculations will be automatically added in the table. From the table it is also possible to export all the data to a spreadsheet by clicking the export icon in the table toolbar.
Relative spectra
It is possible to select a reference spectrum (target spectrum) so that all the other spectra are represented as the relative value to the target. To select the target spectrum click on the icon in the last column. The line will become green.
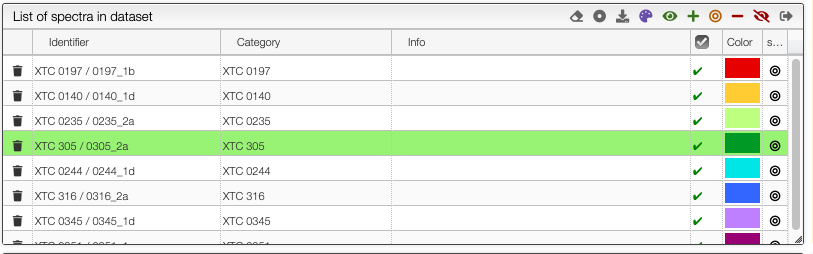
The target spectrum will now be a horizontal line.
In the preferences you can uncheck Relative to come back to the original view.
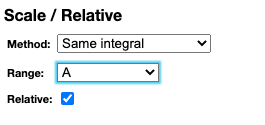
Scaling spectra
Spectra can be rescaled based on the full spectrum or a specific range. Rescale can be based either on the integral, the minimal peak height or maximal peak height.